RNA biology meets developmental biology: post-transcriptional gene regulation in Drosophila development
Development requires that an organism’s genes be expressed at precisely the right time and place, and controlling the transcription of genes to produce mRNAs is just the first step in ensuring that the proteins they encode are only produced when and where they are needed. The localization, translation, and degradation of mRNAs is carefully coordinated to allow the differentiation of specific cell lineages within the developing embryo, as well as the polarization of individual cells to form specialized domains such as neuronal dendrites.
The importance of this post-transcriptional regulation is indicated by the large number of RNA-binding proteins encoded in both the human and Drosophila genomes. RNA-binding proteins and their target RNAs can collaborate to create dynamic subcellular compartments known as granules, where many of the post-transcriptional regulatory steps take place.
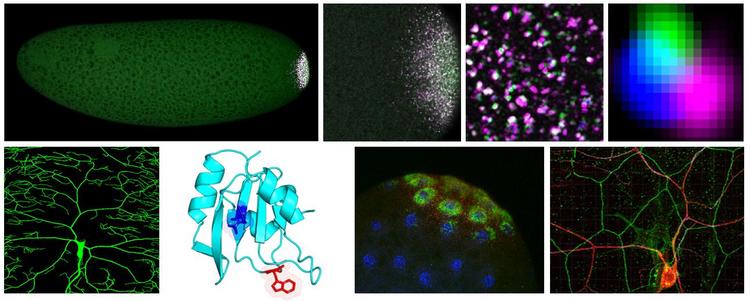
Our lab uses high-resolution microscopy, genetics, and biochemistry to investigate how RNA granules assemble and how they regulate cell fate and function. We have used quantitative single-molecule imaging to examine how different mRNAs become localized to the posterior of the developing Drosophila egg where, due in part to sequences present in the mRNAs themselves, they are packaged into highly organized granules called germ granules. Later, during embryogenesis, the germ granules are segregated to the germ cell progenitors and guide their development to produce the future eggs and sperm.
Recently, we discovered that an RNA-binding protein called Glorund uses a unique mode of interaction to bind and repress the translation of mRNAs that have not been correctly localized to the posterior of the egg. Glorund binds to other RNAs in a different manner and probably regulates their splicing, suggesting that the use of different binding modes may increase the functional versatility of RNA-binding proteins.
We are also interested in how the coordinated regulation of RNA localization and translation controls dendritic arborization in neurons. We have used RNAi and transposon tagging screens to identify numerous RNA-binding proteins and localized mRNAs involved in this process, and have determined how translational repression plays a neuroprotective role to maintain dendritic arbors.
Our current interests include: 1) how and why mRNAs are organized in germ granules; 2) how these granules guide germline development; 3) mechanisms used by RNA-binding proteins to recognize and control translation of their target mRNAs; and 4) roles for RNA-binding proteins in neuronal development.

