2025

Abstract
Germline-soma segregation is crucial for fertility. Primordial germ cells (PGCs) arise early in development and are the very first cells to form in the Drosophila embryo. At the time of PGC formation, the embryo is a syncytium where nuclei divide within a common cytoplasm. Whereas invaginating plasma membrane furrows enclose nuclei to form somatic lineages during the 14th nuclear division cycle, PGCs emerge from the syncytium during the 9th division cycle in a mechanistically distinct process. PGC formation depends on maternally deposited germ granules localized at the embryo's posterior pole. Germ granules trigger protrusion of membrane buds that enlarge to surround several nuclei that reach the posterior pole. Buds are remodeled to cells through mitotic division and constriction of the bud neck. Previous studies implicated F-actin, actin regulators, and contractile ring components in mitotic furrow formation, but what drives bud emergence and how germ granules provoke reshaping of the plasma membrane remain unknown. Here, we investigate the mechanism of germ-granule-induced bud formation. Treating the embryo as a pressurized elastic shell, we used mathematical modeling to examine possible mechanical mechanisms for local membrane protrusion. One mechanism, outward buckling produced by polymerization of a branched F-actin network, is supported by experimental data. Further, we show that germ granules modify membrane lipid composition, promoting local branched F-actin polymerization that initiates PGC formation. We propose that a mechanism for membrane lipid regulation of F-actin dynamics in migrating cells has been adapted for PGC formation in response to spatial cues provided by germ granules.
2024

Abstract
During Drosophila oogenesis, the Oskar (OSK) RNA binding protein (RBP) determines the amount of germ plasm that assembles at the posterior pole of the oocyte. Here, we identify mechanisms that subsequently regulate germ plasm assembly in the early embryo. We show that the Smaug (SMG) RBP is transported into the germ plasm of the early embryo where it accumulates in the germ granules. SMG binds to and represses translation of the messenger RNA (mRNA) as well as the () mRNA, which encodes an RBP that we show promotes germ plasm production. Loss of SMG or mutation of SMG's binding sites in the or mRNA results in excess translation of these transcripts in the germ plasm, accumulation of excess germ plasm, and budding of excess primordial germ cells (PGCs). Therefore, SMG triggers a posttranscriptional regulatory pathway that attenuates the amount of germ plasm in embryos to modulate the number of PGCs.
2023

Abstract
Stem cells in many systems, including germline stem cells (GSCs), increase ribosome biogenesis and translation during terminal differentiation. Here, we show that the H/ACA small nuclear ribonucleoprotein (snRNP) complex that promotes pseudouridylation of ribosomal RNA (rRNA) and ribosome biogenesis is required for oocyte specification. Reducing ribosome levels during differentiation decreased the translation of a subset of messenger RNAs that are enriched for CAG trinucleotide repeats and encode polyglutamine-containing proteins, including differentiation factors such as RNA-binding Fox protein 1. Moreover, ribosomes were enriched at CAG repeats within transcripts during oogenesis. Increasing target of rapamycin (TOR) activity to elevate ribosome levels in H/ACA snRNP complex-depleted germlines suppressed the GSC differentiation defects, whereas germlines treated with the TOR inhibitor rapamycin had reduced levels of polyglutamine-containing proteins. Thus, ribosome biogenesis and ribosome levels can control stem cell differentiation via selective translation of CAG repeat-containing transcripts.

Abstract
Localization of oskar mRNA to the posterior of the Drosophila oocyte is essential for abdominal patterning and germline development. oskar localization is a multi-step process involving temporally and mechanistically distinct transport modes. Numerous cis-acting elements and trans-acting factors have been identified that mediate earlier motor-dependent transport steps leading to an initial accumulation of oskar at the posterior. Little is known, however, about the requirements for the later localization phase, which depends on cytoplasmic flows and results in the accumulation of large oskar ribonucleoprotein granules, called founder granules, by the end of oogenesis. Using super-resolution microscopy, we show that founder granules are agglomerates of smaller oskar transport particles. In contrast to the earlier kinesin-dependent oskar transport, late-phase localization depends on the sequence as well as on the structure of the spliced oskar localization element (SOLE), but not on the adjacent exon junction complex deposition. Late-phase localization also requires the oskar 3' untranslated region (3' UTR), which targets oskar to founder granules. Together, our results show that 3' UTR-mediated targeting together with SOLE-dependent agglomeration leads to accumulation of oskar in large founder granules at the posterior of the oocyte during late stages of oogenesis. In light of previous work showing that oskar transport particles are solid-like condensates, our findings indicate that founder granules form by a process distinct from that of well-characterized ribonucleoprotein granules like germ granules, P bodies, and stress granules. Additionally, they illustrate how an individual mRNA can be adapted to exploit different localization mechanisms depending on the cellular context.

Abstract
Compartmentalization of RNAs and proteins into membraneless structures called granules is a ubiquitous mechanism for organizing and regulating cohorts of RNAs. Germ granules are ribonucleoprotein (RNP) assemblies required for germline development across the animal kingdom, but their regulatory roles in germ cells are not fully understood. We show that after germ cell specification, Drosophila germ granules enlarge through fusion and this growth is accompanied by a shift in function. Whereas germ granules initially protect their constituent mRNAs from degradation, they subsequently target a subset of these mRNAs for degradation while maintaining protection of others. This functional shift occurs through the recruitment of decapping and degradation factors to the germ granules, which is promoted by decapping activators and renders these structures P body-like. Disrupting either the mRNA protection or degradation function results in germ cell migration defects. Our findings reveal plasticity in germ granule function that allows them to be repurposed at different stages of development to ensure population of the gonad by germ cells. Additionally, these results reveal an unexpected level of functional complexity whereby constituent RNAs within the same granule type can be differentially regulated.

Abstract
The Drosophila melanogaster protein Glorund (Glo) represses nanos (nos) translation and uses its quasi-RNA recognition motifs (qRRMs) to recognize both G-tract and structured UA-rich motifs within the nos translational control element (TCE). We showed previously that each of the three qRRMs is multifunctional, capable of binding to G-tract and UA-rich motifs, yet if and how the qRRMs combine to recognize the nos TCE remained unclear. Here we determined solution structures of a nos TCEI_III RNA containing the G-tract and UA-rich motifs. The RNA structure demonstrated that a single qRRM is physically incapable of recognizing both RNA elements simultaneously. In vivo experiments further indicated that any two qRRMs are sufficient to repress nos translation. We probed interactions of Glo qRRMs with TCEI_III RNA using NMR paramagnetic relaxation experiments. Our in vitro and in vivo data support a model whereby tandem Glo qRRMs are indeed multifunctional and interchangeable for recognition of TCE G-tract or UA-rich motifs. This study illustrates how multiple RNA recognition modules within an RNA-binding protein may combine to diversify the RNAs that are recognized and regulated.
2022
Abstract
Dendritic arbor development is a complex, highly regulated process. Post-transcriptional regulation mediated by RNA-binding proteins plays an important role in neuronal dendrite morphogenesis by delivering on-site, on-demand protein synthesis. Here, we show how the Drosophila fragile X mental retardation protein (FMRP), a conserved RNA-binding protein, limits dendrite branching to ensure proper neuronal function during larval sensory neuron development. FMRP knockdown causes increased dendritic terminal branch growth and a resulting overelaboration defect due, in part, to altered microtubule stability and dynamics. FMRP also controls dendrite outgrowth by regulating the Drosophila profilin homolog chickadee (chic). FMRP colocalizes with chic mRNA in dendritic granules and regulates its dendritic localization and protein expression. Whereas RNA-binding domains KH1 and KH2 are both crucial for FMRP-mediated dendritic regulation, KH2 specifically is required for FMRP granule formation and chic mRNA association, suggesting a link between dendritic FMRP granules and FMRP function in dendrite elaboration. Our studies implicate FMRP-mediated modulation of both the neuronal microtubule and actin cytoskeletons in multidendritic neuronal architecture, and provide molecular insight into FMRP granule formation and its relevance to FMRP function in dendritic patterning.

Abstract
MicroRNAs are enriched in neurons and play important roles in dendritic spine development and synaptic plasticity. MicroRNA activity is controlled by a wide range of RNA-binding proteins. FMRP, a highly conserved RNA-binding protein, has been linked to microRNA-mediated gene regulation in axonal development and dendritic spine formation. FMRP also participates in dendritic arbor morphogenesis, but whether and how microRNAs contribute to its function in this process remains to be elucidated. Here, using Drosophila larval sensory neurons, we show that a FMRP-associated microRNA, miR-276, functions in FMRP-mediated space-filling dendrite morphogenesis. Using EGFP microRNA sensors, we demonstrate that FMRP likely acts by regulating miR-276a RNA targeting rather than by modulating microRNA levels. Supporting this conclusion, miR-276a coimmunoprecipitated with FMRP and this association was dependent on the FMRP KH domains. By testing putative targets of the FMRP-miR-276a regulatory axis, we identified nejire as a FMRP-associated mRNA and, using EGFP reporters, showed that the nejire 3' untranslated region is a target of miR-276a in vivo. Genetic analysis places nejire downstream of the FMRP-miR-276a pathway in regulating dendrite patterning. Together, our findings support a model in which FMRP facilitates miR-276a-mediated control of nejire for proper dendrite space-filling morphology and shed light on microRNA-dependent dendrite developmental pathology of fragile X syndrome.

Abstract
Translational control of maternal mRNAs generates spatial and temporal patterns of protein expression necessary to begin animal development. Translational repression of unlocalized nanos (nos) mRNA in late-stage Drosophila oocytes by the hnRNP F/H homolog, Glorund (Glo), is important for embryonic body patterning. While previous work has suggested that repression occurs at both the translation initiation and elongation phases, the molecular mechanism by which Glo regulates nos translation remains elusive. Here, we have identified the Drosophila fragile X mental retardation protein, dFMRP, as a Glo interaction partner with links to the translational machinery. Using an oocyte-based in vitro translation system, we confirmed that Glo regulates both initiation and elongation of a nos translational reporter and showed that dFMRP specifically represses translation elongation and promotes ribosome stalling. Furthermore, we combined mutational analysis and in vivo and in vitro binding assays to show that Glo's qRRM2 domain specifically and directly interacts with dFMRP. Our findings suggest that Glo regulates nos translation elongation by recruiting dFMRP and that Glo's RNA-binding domains can also function as protein-protein interaction interfaces critical for its regulatory functions. Additionally, they reveal a mechanism for targeting dFMRP to specific transcripts.

Abstract
The packaging of specific mRNAs into ribonucleoprotein granules called germ granules is required for germline proliferation and maintenance. During Drosophila germ granule development, mRNAs such as nanos (nos) and polar granule component (pgc) localize to germ granules through a stochastic seeding and self-recruitment process that generates homotypic clusters: aggregates containing multiple copies of a specific transcript. Germ granules vary in mRNA composition with respect to the different transcripts that they contain and their quantity. However, what influences germ granule mRNA composition during development is unclear. To gain insight into how germ granule mRNA heterogeneity arises, we created a computational model that simulates granule development. Although the model includes known mechanisms that were converted into mathematical representations, additional unreported mechanisms proved to be essential for modeling germ granule formation. The model was validated by predicting defects caused by changes in mRNA and protein abundance. Broader application of the model was demonstrated by quantifying nos and pgc localization efficacies and the contribution that an element within the nos 3' untranslated region has on clustering. For the first time, a mathematical representation of Drosophila germ granule formation is described, offering quantitative insight into how mRNA compositions arise while providing a new tool for guiding future studies.
2021
Abstract
Developing oocytes need large supplies of macromolecules and organelles. A conserved strategy for accumulating these products is to pool resources of oocyte-associated germline nurse cells. In Drosophila, these cells grow more than 100-fold to boost their biosynthetic capacity. No previously known mechanism explains how nurse cells coordinate growth collectively. Here, we report a cell cycle-regulating mechanism that depends on bidirectional communication between the oocyte and nurse cells, revealing the oocyte as a critical regulator of germline cyst growth. Transcripts encoding the cyclin-dependent kinase inhibitor, Dacapo, are synthesized by the nurse cells and actively localized to the oocyte. Retrograde movement of the oocyte-synthesized Dacapo protein to the nurse cells generates a network of coupled oscillators that controls the cell cycle of the nurse cells to regulate cyst growth. We propose that bidirectional nurse cell-oocyte communication establishes a growth-sensing feedback mechanism that regulates the quantity of maternal resources loaded into the oocyte.
Abstract
In most animals, the oocyte is the largest cell by volume. The oocyte undergoes a period of large-scale growth during its development, prior to fertilization. At first glance, tissues that support the development of the oocyte in different organisms have diverse cellular characteristics that would seem to prohibit functional comparisons. However, these tissues often act with a common goal of establishing dynamic forms of two-way communication with the oocyte. We propose that this bidirectional communication between oocytes and support cells is a universal phenomenon that can be directly compared across species. Specifically, we highlight fruit fly and mouse oogenesis to demonstrate that similarities and differences in these systems should be used to inform and design future experiments in both models.
2020
Abstract
Dendritic arbor morphology influences how neurons receive and integrate extracellular signals. We show that the ELAV/Hu family RNA-binding protein Found in neurons (Fne) is required for space-filling dendrite growth to generate highly branched arbors of Drosophila larval class IV dendritic arborization neurons. Dendrites of fne mutant neurons are shorter and more dynamic than in wild-type, leading to decreased arbor coverage. These defects result from both a decrease in stable microtubules and loss of dendrite-substrate interactions within the arbor. Identification of transcripts encoding cytoskeletal regulators and cell-cell and cell-ECM interacting proteins as Fne targets using TRIBE further supports these results. Analysis of one target, encoding the cell adhesion protein Basigin, indicates that the cytoskeletal defects contributing to branch instability in fne mutant neurons are due in part to decreased Basigin expression. The ability of Fne to coordinately regulate the cytoskeleton and dendrite-substrate interactions in neurons may shed light on the behavior of cancer cells ectopically expressing ELAV/Hu proteins.

Abstract
Partitioning of mRNAs into ribonucleoprotein (RNP) granules supports diverse regulatory programs within the crowded cytoplasm. At least two types of RNP granules populate the germ plasm, a cytoplasmic domain at the posterior of the oocyte and embryo. Germ granules deliver mRNAs required for germline development to pole cells, the germ cell progenitors. A second type of RNP granule, here named founder granules, contains mRNA, which encodes the germ plasm organizer. Whereas mRNA is essential for germ plasm assembly during oogenesis, we show that it is toxic to pole cells. Founder granules mediate compartmentalized degradation of during embryogenesis to minimize its inheritance by pole cells. Degradation of in founder granules is temporally and mechanistically distinct from degradation of and other mRNAs during the maternal-to-zygotic transition. Our results show how compartmentalization in RNP granules differentially controls fates of mRNAs localized within the same cytoplasmic domain.

Abstract
mRNAs enriched in membraneless condensates provide functional compartmentalization within cells. The mechanisms that recruit transcripts to condensates are under intense study; however, how mRNAs organize once they reach a granule remains poorly understood. Here, we report on a self-sorting mechanism by which multiple mRNAs derived from the same gene assemble into discrete homotypic clusters. We demonstrate that in vivo mRNA localization to granules and self-assembly within granules are governed by different mRNA features: localization is encoded by specific RNA regions, whereas self-assembly involves the entire mRNA, does not involve sequence-specific, ordered intermolecular RNA:RNA interactions, and is thus RNA sequence independent. We propose that the ability of mRNAs to self-sort into homotypic assemblies is an inherent property of an messenger ribonucleoprotein (mRNP) that is augmented under conditions that increase RNA concentration, such as upon enrichment in RNA-protein granules, a process that appears conserved in diverse cellular contexts and organisms.
2018
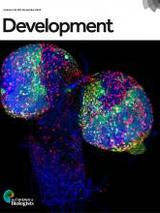
Abstract
Specification and development of germ cells depend on molecular determinants within the germ plasm, a specialized cytoplasmic domain at the posterior of the embryo. Localization of numerous mRNAs to the germ plasm occurs by their incorporation, as single-transcript ribonucleoprotein (RNP) particles, into complex RNP granules called polar granules. Incorporation of mRNAs into polar granules is followed by recruitment of additional like transcripts to form discrete homotypic clusters. The -acting localization signals that target mRNAs to polar granules and promote homotypic clustering remain largely uncharacterized. Here, we show that the () and () 3' untranslated regions contain complex localization signals comprising multiple, independently weak and partially functionally redundant localization elements (LEs). We demonstrate that targeting of to polar granules and self-assembly into homotypic clusters are functionally separable processes mediated by distinct classes of LEs. We identify a sequence motif shared by other polar granule mRNAs that contributes to homotypic clustering. Our results suggest that mRNA localization signal complexity may be a feature required by the targeting and self-recruitment mechanism that drives germ plasm mRNA localization.

Abstract
The formation of ribonucleoprotein assemblies called germ granules is a conserved feature of germline development. In Drosophila, germ granules form at the posterior of the oocyte in a specialized cytoplasm called the germ plasm, which specifies germline fate during embryogenesis. mRNAs, including nanos (nos) and polar granule component (pgc), that function in germline development are localized to the germ plasm through their incorporation into germ granules, which deliver them to the primordial germ cells. Germ granules are nucleated by Oskar (Osk) protein and contain varying combinations and quantities of their constituent mRNAs, which are organized as spatially distinct, multi-copy homotypic clusters. The process that gives rise to such heterogeneous yet organized granules remains unknown. Here, we show that individual nos and pgc transcripts can populate the same nascent granule, and these first transcripts then act as seeds, recruiting additional like transcripts to form homotypic clusters. Within a granule, homotypic clusters grow independently of each other but depend on the simultaneous acquisition of additional Osk. Although granules can contain multiple clusters of a particular mRNA, granule mRNA content is dominated by cluster size. These results suggest that the accumulation of mRNAs in the germ plasm is controlled by the mRNAs themselves through their ability to form homotypic clusters; thus, RNA self-association drives germ granule mRNA localization. We propose that a stochastic seeding and self-recruitment mechanism enables granules to simultaneously incorporate many different mRNAs while ensuring that each becomes enriched to a functional threshold.
2017

Abstract
Most cellular stresses induce protein translation inhibition and stress granule formation. Here, using Drosophila S2 cells, we investigate the role of G3BP/Rasputin in this process. In contrast to arsenite treatment, where dephosphorylated Ser142 Rasputin is recruited to stress granules, we find that, upon amino acid starvation, only the phosphorylated Ser142 form is recruited. Furthermore, we identify Sec16, a component of the endoplasmic reticulum exit site, as a Rasputin interactor and stabilizer. Sec16 depletion results in Rasputin degradation and inhibition of stress granule formation. However, in the absence of Sec16, pharmacological stabilization of Rasputin is not enough to rescue the assembly of stress granules. This is because Sec16 specifically interacts with phosphorylated Ser142 Rasputin, the form required for stress granule formation upon amino acid starvation. Taken together, these results demonstrate that stress granule formation is fine-tuned by specific signaling cues that are unique to each stress. These results also expand the role of Sec16 as a stress response protein.

Abstract
The primordial germ cells (PGCs) specified during embryogenesis serve as progenitors to the adult germline stem cells. In Drosophila, the proper specification and formation of PGCs require both centrosomes and germ plasm, which contains the germline determinants. Centrosomes are microtubule (MT)-organizing centers that ensure the faithful segregation of germ plasm into PGCs. To date, mechanisms that modulate centrosome behavior to engineer PGC development have remained elusive. Only one germ plasm component, Germ cell-less (Gcl), is known to play a role in PGC formation. Here, we show that Gcl engineers PGC formation by regulating centrosome dynamics. Loss of gcl leads to aberrant centrosome separation and elaboration of the astral MT network, resulting in inefficient germ plasm segregation and aborted PGC cellularization. Importantly, compromising centrosome separation alone is sufficient to mimic the gcl loss-of-function phenotypes. We conclude Gcl functions as a key regulator of centrosome separation required for proper PGC development.

Abstract
The Drosophila hnRNP F/H homolog, Glorund (Glo), regulates nanos mRNA translation by interacting with a structured UA-rich motif in the nanos 3' untranslated region. Glo regulates additional RNAs, however, and mammalian homologs bind G-tract sequences to regulate alternative splicing, suggesting that Glo also recognizes G-tract RNA. To gain insight into how Glo recognizes both structured UA-rich and G-tract RNAs, we used mutational analysis guided by crystal structures of Glo's RNA-binding domains and identified two discrete RNA-binding surfaces that allow Glo to recognize both RNA motifs. By engineering Glo variants that favor a single RNA-binding mode, we show that a subset of Glo's functions in vivo is mediated solely by the G-tract binding mode, whereas regulation of nanos requires both recognition modes. Our findings suggest a molecular mechanism for the evolution of dual RNA motif recognition in Glo that may be applied to understanding the functional diversity of other RNA-binding proteins.

Abstract
Spatial arrangement of different neuron types within a territory is essential to neuronal development and function. How development of different neuron types is coordinated for spatial coexistence is poorly understood. In Drosophila, dendrites of four classes of dendritic arborization (C1-C4da) neurons innervate overlapping receptive fields within the larval epidermis. These dendrites are intermittently enclosed by epidermal cells, with different classes exhibiting varying degrees of enclosure. The role of enclosure in neuronal development and its underlying mechanism remain unknown. We show that the membrane-associated protein Coracle acts in C4da neurons and epidermal cells to locally restrict dendrite branching and outgrowth by promoting enclosure. Loss of C4da neuron enclosure results in excessive branching and growth of C4da neuron dendrites and retraction of C1da neuron dendrites due to local inhibitory interactions between neurons. We propose that enclosure of dendrites by epidermal cells is a developmental mechanism for coordinated innervation of shared receptive fields.
2016

Abstract
The ability to visualize RNA in situ is essential to dissect mechanisms for the temporal and spatial regulation of gene expression that drives development. Although considerable attention has been focused on transcriptional control, studies in model organisms like Drosophila have highlighted the importance of post-transcriptional mechanisms - most notably intracellular mRNA localization - in the formation and patterning of the body axes, specification of cell fates, and polarized cell functions. Our understanding of both types of regulation has been greatly advanced by technological innovations that enable a combination of highly quantitative and dynamic analysis of RNA. This review presents two methods, single molecule fluorescence in situ hybridization for high resolution quantitative RNA detection in fixed Drosophila oocytes and embryos and genetically encoded fluorescent RNA labeling for detection in live cells.

Abstract
Dendritic arbor morphology is a key determinant of neuronal function. Once established, dendrite branching patterns must be maintained as the animal develops to ensure receptive field coverage. The translational repressors Nanos (Nos) and Pumilio (Pum) are required to maintain dendrite growth and branching of Drosophila larval class IV dendritic arborization (da) neurons, but their specific regulatory role remains unknown. We show that Nos-Pum-mediated repression of the pro-apoptotic gene head involution defective (hid) is required to maintain a balance of dendritic growth and retraction in class IV da neurons and that upregulation of hid results in decreased branching because of an increase in caspase activity. The temporal requirement for nos correlates with an ecdysone-triggered switch in sensitivity to apoptotic stimuli that occurs during the mid-L3 transition. We find that hid is required during pupariation for caspase-dependent pruning of class IV da neurons and that Nos and Pum delay pruning. Together, these results suggest that Nos and Pum provide a crucial neuroprotective regulatory layer to ensure that neurons behave appropriately in response to developmental cues.

Abstract
Localizing messenger RNAs at specific subcellular sites is a conserved mechanism for targeting the synthesis of cytoplasmic proteins to distinct subcellular domains, thereby generating the asymmetric protein distributions necessary for cellular and developmental polarity. However, the full range of transcripts that are asymmetrically distributed in specialized cell types, and the significance of their localization, especially in the nervous system, are not known. We used the EP-MS2 method, which combines EP transposon insertion with the MS2/MCP in vivo fluorescent labeling system, to screen for novel localized transcripts in polarized cells, focusing on the highly branched Drosophila class IV dendritic arborization neurons. Of a total of 541 lines screened, we identified 55 EP-MS2 insertions producing transcripts that were enriched in neuronal processes, particularly in dendrites. The 47 genes identified by these insertions encode molecularly diverse proteins, and are enriched for genes that function in neuronal development and physiology. RNAi-mediated knockdown confirmed roles for many of the candidate genes in dendrite morphogenesis. We propose that the transport of mRNAs encoded by these genes into the dendrites allows their expression to be regulated on a local scale during the dynamic developmental processes of dendrite outgrowth, branching, and/or remodeling.

Abstract
Drosophila larval dendritic arborization (da) neurons are a popular model for investigating mechanisms of neuronal morphogenesis. Da neurons develop in communication with the epidermal cells they innervate and thus their analysis benefits from in situ visualization of both neuronally and epidermally expressed proteins by immunofluorescence. Traditional methods of preparing larval fillets for immunofluorescence experiments leave intact the muscle tissue that covers most of the body wall, presenting several challenges to imaging neuronal and epidermal proteins. Here we describe a method for removing muscle tissue from Drosophila larval fillets. This protocol enables imaging of proteins that are otherwise obscured by muscle tissue, improves signal to noise ratio, and facilitates the use of super-resolution microscopy to study da neuron development.

Abstract
bicoid mRNA localises to the Drosophila oocyte anterior from stage 9 of oogenesis onwards to provide a local source for Bicoid protein for embryonic patterning. Live imaging at stage 9 reveals that bicoid mRNA particles undergo rapid Dynein-dependent movements near the oocyte anterior, but with no directional bias. Furthermore, bicoid mRNA localises normally in shot(2A2), which abolishes the polarised microtubule organisation. FRAP and photo-conversion experiments demonstrate that the RNA is stably anchored at the anterior, independently of microtubules. Thus, bicoid mRNA is localised by random active transport and anterior anchoring. Super-resolution imaging reveals that bicoid mRNA forms 110-120 nm particles with variable RNA content, but constant size. These particles appear to be well-defined structures that package the RNA for transport and anchoring.
2015

Abstract
Messenger RNA localization is a conserved mechanism for spatial control of protein synthesis, with key roles in generating cellular and developmental asymmetry. Whereas different transcripts may be targeted to the same subcellular domain, the extent to which their localization is coordinated is unclear. Using quantitative single-molecule imaging, we analysed the assembly of Drosophila germ plasm mRNA granules inherited by nascent germ cells. We find that the germ-cell-destined transcripts nanos, cyclin B and polar granule component travel within the oocyte as ribonucleoprotein particles containing single mRNA molecules but co-assemble into multi-copy heterogeneous granules selectively at the posterior of the oocyte. The stoichiometry and dynamics of assembly indicate a defined stepwise sequence. Our data suggest that co-packaging of these transcripts ensures their effective segregation to germ cells. In contrast, compartmentalization of the germline determinant oskar mRNA into different granules limits its entry into germ cells. This exclusion is required for proper germline development.

Abstract
Drosophila telomeres constitute a remarkable exception to the telomerase mechanism. Although maintaining the same cytological and functional properties as telomerase maintain telomeres, Drosophila telomeres embed the telomere retrotransposons whose specific and highly regulated terminal transposition maintains the appropriate telomere length in this organism. Nevertheless, our current understanding of how the mechanism of the retrotransposon telomere works and which features are shared with the telomerase system is very limited. We report for the first time a detailed study of the localization of the main components that constitute the telomeres in Drosophila, HeT-A and TART RNAs and proteins. Our results in wild type and mutant strains reveal localizations of HeT-A Gag and TART Pol that give insight in the behavior of the telomere retrotransposons and their control. We find that TART Pol and HeT-A Gag only co-localize at the telomeres during the interphase of cells undergoing mitotic cycles. In addition, unexpected protein and RNA localizations with a well-defined pattern in cells such as the ovarian border cells and nurse cells, suggest possible strategies for the telomere transposons to reach the oocyte, and/or additional functions that might be important for the correct development of the organism. Finally, we have been able to visualize the telomere RNAs at different ovarian stages of development in wild type and mutant lines, demonstrating their presence in spite of being tightly regulated by the piRNA mechanism.
2014

Abstract
The large number of RNA-binding proteins and translation factors encoded in the Drosophila and other metazoan genomes predicts widespread use of post-transcriptional regulation in cellular and developmental processes. Previous studies identified roles for several RNA-binding proteins in dendrite branching morphogenesis of Drosophila larval sensory neurons. To determine the larger contribution of post-transcriptional gene regulation to neuronal morphogenesis, we conducted an RNA interference screen to identify additional Drosophila proteins annotated as either RNA-binding proteins or translation factors that function in producing the complex dendritic trees of larval class IV dendritic arborization neurons. We identified 88 genes encoding such proteins whose knockdown resulted in aberrant dendritic morphology, including alterations in dendritic branch number, branch length, field size, and patterning of the dendritic tree. In particular, splicing and translation initiation factors were associated with distinct and characteristic phenotypes, suggesting that different morphogenetic events are best controlled at specific steps in post-transcriptional messenger RNA metabolism. Many of the factors identified in the screen have been implicated in controlling the subcellular distributions and translation of maternal messenger RNAs; thus, common post-transcriptional regulatory strategies may be used in neurogenesis and in the generation of asymmetry in the female germline and embryo.
2013

Abstract
Ribosomes can read through stop codons in a regulated manner, elongating rather than terminating the nascent peptide. Stop codon readthrough is essential to diverse viruses, and phylogenetically predicted to occur in a few hundred genes in Drosophila melanogaster, but the importance of regulated readthrough in eukaryotes remains largely unexplored. Here, we present a ribosome profiling assay (deep sequencing of ribosome-protected mRNA fragments) for Drosophila melanogaster, and provide the first genome-wide experimental analysis of readthrough. Readthrough is far more pervasive than expected: the vast majority of readthrough events evolved within D. melanogaster and were not predicted phylogenetically. The resulting C-terminal protein extensions show evidence of selection, contain functional subcellular localization signals, and their readthrough is regulated, arguing for their importance. We further demonstrate that readthrough occurs in yeast and humans. Readthrough thus provides general mechanisms both to regulate gene expression and function, and to add plasticity to the proteome during evolution. DOI: http://dx.doi.org/10.7554/eLife.01179.001.

Abstract
Localized cytoplasmic determinants packaged as ribonucleoprotein (RNP) particles direct embryonic patterning and cell fate specification in a wide range of organisms. Once established, the asymmetric distributions of such RNP particles must be maintained, often over considerable developmental time. A striking example is the Drosophila germ plasm, which contains RNP particles whose localization to the posterior of the egg during oogenesis results in their asymmetric inheritance and segregation of germline from somatic fates in the embryo. Although actin-based anchoring mechanisms have been implicated, high-resolution live imaging revealed persistent trafficking of germ plasm RNP particles at the posterior cortex of the Drosophila oocyte. This motility relies on cortical microtubules, is mediated by kinesin and dynein motors, and requires coordination between the microtubule and actin cytoskeletons. Finally, we show that RNP particle motility is required for long-term germ plasm retention. We propose that anchoring is a dynamic state that renders asymmetries robust to developmental time and environmental perturbations.

Abstract
Stochastic mechanisms are sometimes utilized to diversify cell fates, especially in nervous systems. In the Drosophila retina, stochastic expression of the PAS-bHLH transcription factor Spineless (Ss) controls photoreceptor subtype choice. In one randomly distributed subset of R7 photoreceptors, Ss activates Rhodopsin4 (Rh4) and represses Rhodopsin3 (Rh3); counterparts lacking Ss express Rh3 and repress Rh4. In the dorsal third region of the retina, the Iroquois Complex transcription factors induce Rh3 in Rh4-expressing R7s. Here, we show that Ss levels are controlled in a binary on/off manner throughout the retina yet are attenuated in the dorsal third region to allow Rh3 coexpression with Rh4. Whereas the sensitivity of rh3 repression to differences in Ss levels generates stochastic and regionalized patterns, the robustness of rh4 activation ensures its stochastic expression throughout the retina. Our findings show how stochastic and regional inputs are integrated to control photoreceptor subtype specification in the Drosophila retina.
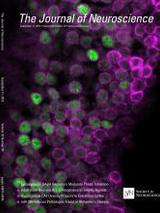
Abstract
Intracellular mRNA localization is a conserved mechanism for spatially regulating protein production in polarized cells, such as neurons. The mRNA encoding the translational repressor Nanos (Nos) forms ribonucleoprotein (RNP) particles that are dendritically localized in Drosophila larval class IV dendritic arborization (da) neurons. In nos mutants, class IV da neurons exhibit reduced dendritic branching complexity, which is rescued by transgenic expression of wild-type nos mRNA but not by a localization-compromised nos derivative. While localization is essential for nos function in dendrite morphogenesis, the mechanism underlying the transport of nos RNP particles was unknown. We investigated the mechanism of dendritic nos mRNA localization by analyzing requirements for nos RNP particle motility in class IV da neuron dendrites through live imaging of fluorescently labeled nos mRNA. We show that dynein motor machinery components mediate transport of nos mRNA in proximal dendrites. Two factors, the RNA-binding protein Rumpelstiltskin and the germ plasm protein Oskar, which are required for diffusion/entrapment-mediated localization of nos during oogenesis, also function in da neurons for formation and transport of nos RNP particles. Additionally, we show that nos regulates neuronal function, most likely independent of its dendritic localization and function in morphogenesis. Our results reveal adaptability of localization factors for regulation of a target transcript in different cellular contexts.
2012

Abstract
The translational regulators Nanos (Nos) and Pumilio (Pum) work together to regulate the morphogenesis of dendritic arborization (da) neurons of the Drosophila larval peripheral nervous system. In contrast, Nos and Pum function in opposition to one another in the neuromuscular junction to regulate the morphogenesis and the electrophysiological properties of synaptic boutons. Neither the cellular functions of Nos and Pum nor their regulatory targets in neuronal morphogenesis are known. Here we show that Nos and Pum are required to maintain the dendritic complexity of da neurons during larval growth by promoting the outgrowth of new dendritic branches and the stabilization of existing dendritic branches, in part by regulating the expression of cut and head involution defective. Through an RNA interference screen we uncover a role for the translational co-factor Brain Tumor (Brat) in dendrite morphogenesis of da neurons and demonstrate that Nos, Pum, and Brat interact genetically to regulate dendrite morphogenesis. In the neuromuscular junction, Brat function is most likely specific for Pum in the presynaptic regulation of bouton morphogenesis. Our results reveal how the combinatorial use of co-regulators like Nos, Pum and Brat can diversify their roles in post-transcriptional regulation of gene expression for neuronal morphogenesis.
2011

Abstract
Translational control of gene expression is essential for development in organisms that rely on maternal mRNAs. In Drosophila, translation of maternal nanos (nos) mRNA must be restricted to the posterior of the early embryo for proper patterning of the anterior-posterior axis. Spatial control of nos translation is coordinated through the localization of a small subset of nos mRNA to the posterior pole late in oogenesis, activation of this localized mRNA, and repression of the remaining unlocalized nos mRNA throughout the bulk cytoplasm. Translational repression is mediated by the interaction of a cis-acting element in the nos 3' untranslated region with two proteins, Glorund (Glo) and Smaug (Smg), that function in the oocyte and embryo, respectively. The mechanism of Glo-dependent repression is unknown. Previous work suggests that Smg represses translation initiation but this model is not easily reconciled with evidence for polysome association of repressed nos mRNA. Using an in vitro translation system, we have decoupled translational repression of nos imposed during oogenesis from repression during embryogenesis. Our results suggest that both Glo and Smg regulate translation initiation, but by different mechanisms. Furthermore, we show that, during late oogenesis, nos translation is also repressed post-initiation and provide evidence that Glo mediates this event. This post-initiation block is maintained into embryogenesis during the transition to Smg-dependent regulation. We propose that the use of multiple modes of repression ensures inactivation of nos RNA that is translated at earlier stages of oogenesis and maintenance of this inactivate state throughout late oogenesis into embryogenesis.

Abstract
Localization of nanos (nos) mRNA to the posterior pole of the Drosophila oocyte is essential for abdominal segmentation and germline development during embryogenesis. Posterior localization is mediated by a complex cis-acting localization signal in the nos 3' untranslated region that comprises multiple partially redundant elements. Genetic analysis suggests that this signal is recognized by RNA-binding proteins and associated factors that package nos mRNA into a localization competent ribonucleoprotein complex. However, functional redundancy among localization elements has made the identification of individual localization factors difficult. Indeed, only a single direct-acting nos localization factor, Rumpelstiltskin (Rump), has been identified thus far. Through a sensitized genetic screen, we have now identified the Argonaute family member Aubergine (Aub) as a nos localization factor. Aub interacts with nos mRNA in vivo and co-purifies with Rump in an RNA-dependent manner. Our results support a role for Aub, independent of its function in RNA silencing, as a component of a nos mRNA localization complex.

Abstract
Many RNAs show polarized or otherwise non-random subcellular distributions. To create a method for genome-wide genetic screens for RNAs with asymmetric subcellular distributions, we have combined methods for gene tagging and live imaging of messenger RNA (mRNA). A pilot screen in a highly polarized, differentiated cell in the Drosophila larva, the branched terminal cell of the tracheal system, demonstrates the feasibility of the method for identifying new asymmetrically localized mRNAs in vivo.
Abstract
BACKGROUND: In many organisms, germ cells are segregated from the soma through the inheritance of the specialized germ plasm, which contains mRNAs and proteins that specify germ cell fate and promote germline development. Whereas germ plasm assembly has been well characterized, mechanisms mediating germ plasm inheritance are poorly understood. In the Drosophila embryo, germ plasm is anchored to the posterior cortex, and nuclei that migrate into this region give rise to the germ cell progenitors, or pole cells. How the germ plasm interacts with these nuclei for pole cell induction and is selectively incorporated into the forming pole cells is not known. RESULTS: Live imaging of two conserved germ plasm components, nanos mRNA and Vasa protein, revealed that germ plasm segregation is a dynamic process involving active transport of germ plasm RNA-protein complexes coordinated with nuclear migration. We show that centrosomes accompanying posterior nuclei induce release of germ plasm from the cortex and recruit these components by dynein-dependent transport on centrosome-nucleated microtubules. As nuclei divide, continued transport on astral microtubules partitions germ plasm to daughter nuclei, leading to its segregation into pole cells. Disruption of these transport events prevents incorporation of germ plasm into pole cells and impairs germ cell development. CONCLUSIONS: Our results indicate that active transport of germ plasm is essential for its inheritance and ensures the production of a discrete population of germ cell progenitors endowed with requisite factors for germline development. Transport on astral microtubules may provide a general mechanism for the segregation of cell fate determinants.
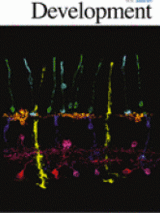
Abstract
Asymmetric mRNA localization is an effective mechanism for establishing cellular and developmental polarity. Posterior localization of oskar in the Drosophila oocyte targets the synthesis of Oskar to the posterior, where Oskar initiates the assembly of the germ plasm. In addition to harboring germline determinants, the germ plasm is required for localization and translation of the abdominal determinant nanos. Consequently, failure of oskar localization during oogenesis results in embryos lacking germ cells and abdominal segments. oskar accumulates at the oocyte posterior during mid-oogenesis through a well-studied process involving kinesin-mediated transport. Through live imaging of oskar mRNA, we have uncovered a second, mechanistically distinct phase of oskar localization that occurs during late oogenesis and results in amplification of the germ plasm. Analysis of two newly identified oskar localization factors, Rumpelstiltskin and Lost, that are required specifically for this late phase of oskar localization shows that germ plasm amplification ensures robust abdomen and germ cell formation during embryogenesis. In addition, our results indicate the importance of mechanisms for adapting mRNAs to utilize multiple localization pathways as necessitated by the dramatic changes in ovarian physiology that occur during oogenesis.
2010

Abstract
Localization of the germ plasm to the posterior of the Drosophila oocyte is required for anteroposterior patterning and germ cell development during embryogenesis. While mechanisms governing the localization of individual germ plasm components have been elucidated, the process by which germ plasm assembly is restricted to the posterior pole is poorly understood. In this study, we identify a novel allele of bazooka (baz), the Drosophila homolog of Par-3, which has allowed the analysis of baz function throughout oogenesis. We demonstrate that baz is required for spatial restriction of the germ plasm and axis patterning, and we uncover multiple requirements for baz in regulating the organization of the oocyte microtubule cytoskeleton. Our results suggest that distinct cortical domains established by Par proteins polarize the oocyte through differential effects on microtubule organization. We further show that microtubule plus-end enrichment is sufficient to drive germ plasm assembly even at a distance from the oocyte cortex, suggesting that control of microtubule organization is critical not only for the localization of germ plasm components to the posterior of the oocyte but also for the restriction of germ plasm assembly to the posterior pole.
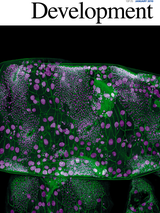
Abstract
Localization of bicoid mRNA to the anterior of the Drosophila oocyte is essential for patterning the anteroposterior body axis in the early embryo. bicoid mRNA localizes in a complex multistep process involving transacting factors, molecular motors and cytoskeletal components that remodel extensively during the lifetime of the mRNA. Genetic requirements for several localization factors, including Swallow and Staufen, are well established, but the precise roles of these factors and their relationship to bicoid mRNA transport particles remains unresolved. Here we use live cell imaging, super-resolution microscopy in fixed cells and immunoelectron microscopy on ultrathin frozen sections to study the distribution of Swallow, Staufen, actin and dynein relative to bicoid mRNA during late oogenesis. We show that Swallow and bicoid mRNA are transported independently and are not colocalized at their final destination. Furthermore, Swallow is not required for bicoid transport. Instead, Swallow localizes to the oocyte plasma membrane, in close proximity to actin filaments, and we present evidence that Swallow functions during the late phase of bicoid localization by regulating the actin cytoskeleton. In contrast, Staufen, dynein and bicoid mRNA form nonmembranous, electron dense particles at the oocyte anterior. Our results exclude a role for Swallow in linking bicoid mRNA to the dynein motor. Instead we propose a model for bicoid mRNA localization in which Swallow is transported independently by dynein and contributes indirectly to bicoid mRNA localization by organizing the cytoskeleton, whereas Staufen plays a direct role in dynein-dependent bicoid mRNA transport.
2009
Abstract
The asymmetric localization of four maternal mRNAs - gurken, bicoid, oskar and nanos - in the Drosophila oocyte is essential for the development of the embryonic body axes. Fluorescent imaging methods are now being used to visualize these mRNAs in living tissue, allowing dynamic analysis of their behaviors throughout the process of localization. This review summarizes recent findings from such studies that provide new insight into the elaborate cellular mechanisms that are used to transport mRNAs to different regions of the oocyte and to maintain their localized distributions during oogenesis.

Abstract
Precise temporal and spatial regulation of gene expression during Drosophila oogenesis is essential for patterning the anterior-posterior and dorsal-ventral body axes. Establishment of the anterior-posterior axis requires posterior localization and translational control of both oskar and nanos mRNAs. Establishment of the dorsal-ventral axis depends on the precise restriction of gurken mRNA and protein to the dorsal-anterior corner of the oocyte. We have previously shown that Glorund, the Drosophila hnRNP F/H homolog, contributes to anterior-posterior axis patterning by regulating translation of nanos mRNA, through a direct interaction with its 3' untranslated region. To investigate the pleiotropy of the glorund mutant phenotype, which includes dorsal-ventral and nuclear morphology defects, we searched for proteins that interact with Glorund. Here we show that Glorund is part of a complex containing the hnRNP protein Hrp48 and the splicing factor Half-pint and plays a role both in mRNA localization and nurse cell chromosome organization, probably by regulating alternative splicing of ovarian tumor. We propose that Glorund is a component of multiple protein complexes and functions both as a translational repressor and splicing regulator for anterior-posterior and dorsal-ventral patterning.
Abstract
Pumilio (Pum) is a translational repressor that binds selectively to target mRNAs and recruits Nanos (Nos) as a corepressor. In the larval neuromuscular system, Pum represses expression of the translation factor eIF-4E and the glutamate receptor subunit GluRIIA. Here, we show that Nos, like Pum, is expressed at the neuromuscular junction (NMJ) and in neuronal cell bodies. Surprisingly, however, Nos and Pum have divergent functions on both the presynaptic and postsynaptic sides of the NMJ. In nos mutant and nos RNA interference larvae, the number of NMJ boutons is increased, whereas loss of Pum reduces the bouton number. On the postsynaptic side, Nos acts in opposition to Pum in regulating the subunit composition of the glutamate receptor. NMJ active zones are associated with GluRIIA- and GluRIIB-containing receptor clusters. Loss of Nos causes downregulation of GluRIIA and increases the levels of GluRIIB. Consistent with this finding, the electrophysiological properties of NMJs lacking postsynaptic Nos suggest that they use primarily GluRIIB-containing receptors. Nos can regulate GluRIIB in the absence of GluRIIA, suggesting that the effects of Nos on GluRIIB levels are at least partially independent of synaptic competition between GluRIIA and GluRIIB. Nos is a target for Pum repression, and Pum binds selectively to the 3' untranslated regions of the nos and GluRIIA mRNAs. Our results suggest a model in which regulatory interplay among Pum, Nos, GluRIIA, and GluRIIB could cause a small change in Pum activity to be amplified into a large shift in the balance between GluRIIA and GluRIIB synapses.
2008

Abstract
Spatial control of mRNA translation can generate cellular asymmetries and functional specialization of polarized cells like neurons. A requirement for the translational repressor Nanos (Nos) in the Drosophila larval peripheral nervous system (PNS) implicates translational control in dendrite morphogenesis [1]. Nos was first identified by its requirement in the posterior of the early embryo for abdomen formation [2]. Nos synthesis is targeted to the posterior pole of the oocyte and early embryo through translational repression of unlocalized nos mRNA coupled with translational activation of nos mRNA localized at the posterior pole [3, 4]. Abolishment of nos localization prevents abdominal development, whereas translational derepression of unlocalized nos mRNA suppresses head/thorax development, emphasizing the importance of spatial regulation of nos mRNA [3, 5]. Loss and overexpression of Nos affect dendrite branching complexity in class IV dendritic arborization (da) neurons, suggesting that nos also might be regulated in these larval sensory neurons [1]. Here, we show that localization and translational control of nos mRNA are essential for da neuron morphogenesis. RNA-protein interactions that regulate nos translation in the oocyte and early embryo also regulate nos in the PNS. Live imaging of nos mRNA shows that the cis-acting signal responsible for posterior localization in the oocyte/embryo mediates localization to the processes of class IV da neurons but suggests a different transport mechanism. Targeting of nos mRNA to the processes of da neurons may reflect a local requirement for Nos protein in dendritic translational control.

Abstract
The development of a functional germline is essential for species propagation. The nanos (nos) gene plays an evolutionarily conserved role in germline development and is also essential for abdominal patterning in Drosophila. A small fraction of nos mRNA is localized to the germ plasm at the posterior pole of the Drosophila embryo, where it becomes incorporated into the germ cells. Germ plasm associated nos mRNA is translated to produce a gradient of Nos protein that patterns the abdomen, whereas the remaining unlocalized RNA is translationally repressed to allow anterior development. Using transgenes that compromise nos mRNA localization and translational regulation, we show that wild-type body patterning can ensue without nos mRNA localization provided that nos translation is properly modulated. In contrast, localization of nos to the germ plasm, but not translational regulation, is essential for nos function in the developing germ cells. We propose that an imperative for nos localization in producing a functional germline has preserved an inefficient localization mechanism.
Abstract
Anterior-posterior axis patterning of the Drosophila embryo requires Nanos activity selectively in the posterior. This spatial asymmetry of Nanos is generated by the localization of nanos mRNA to the posterior pole of the embryo, where it is subsequently translated. Posterior localization of nanos is mediated by a complex cis-acting localization signal in its 3' untranslated region comprising several partially redundant localization elements. This localization signal redundancy has hampered the identification of trans-acting factors that act specifically to effect posterior localization of nanos. Here, we have used a biochemical approach to identify Rumpelstiltskin, a Drosophila heterogeneous nuclear ribonucleoprotein (hnRNP) M homolog, which binds directly to an individual nanos localization element. Rumpelstiltskin associates with nanos mRNA in vitro and in vivo, and binding by Rumpelstiltskin correlates with localization element function in vivo. Through analysis of a rumpelstiltskin null mutation by genetic strategies that circumvent redundancy, we demonstrate that Rumpelstiltskin regulates anterior-posterior axis patterning by functioning as a direct-acting nanos mRNA localization factor.
Abstract
During Drosophila oogenesis, the targeted localization of gurken (grk) mRNA leads to the establishment of the axis polarity of the egg. In early stages of oogenesis, grk mRNA is found at the posterior of the oocyte, whereas in the later stages grk mRNA is positioned at the dorsal anterior corner of the oocyte. In order to visualize the real-time localization and anchorage of endogenous grk mRNA in living oocytes, we have utilized the MS2-MCP system. We show that MCP-GFP-tagged endogenous grk mRNA localizes properly within wild-type oocytes and behaves aberrantly in mutant backgrounds. Fluorescence recovery after photobleaching (FRAP) experiments of localized grk mRNA in egg chambers reveal a difference in the dynamics of grk mRNA between young and older egg chambers. grk mRNA particles, as a population, are highly dynamic molecules that steadily lose their dynamic nature as oogenesis progresses. This difference in dynamics is attenuated in K10 and sqd(1) mutants such that mislocalized grk mRNA in older stages is much more dynamic compared with that in wild-type controls. By contrast, in flies with compromised dynein activity, properly localized grk mRNA is much more static. Taken together, we have observed the nature of localized grk mRNA in live oocytes and propose that its maintenance changes from a dynamic to a static process as oogenesis progresses.

Abstract
Intracellular mRNA localization directs protein synthesis to particular subcellular domains to establish embryonic polarity in a variety of organisms. In Drosophila, bicoid (bcd) mRNA is prelocalized at the oocyte anterior. After fertilization, translation of this RNA produces a Bcd protein gradient that determines anterior cell fates [1] and [2]. Analysis of bcd mRNA during late stages of oogenesis suggested a model for steady-state bcd localization by continual active transport [3]. However, this mechanism cannot explain maintenance of bcd localization throughout the end of oogenesis, when microtubules disassemble in preparation for embryogenesis [4] and [5], or retention of bcd at the anterior in mature oocytes, which can remain dormant for weeks before fertilization [6]. Here, we elucidate the path and mechanism of sustained bcd mRNA transport by direct observation of bcd RNA particle translocation in living oocytes. We show that bcd mRNA shifts from continuous active transport to stable actin-dependent anchoring at the end of oogenesis. Egg activation triggers bcd release from the anterior cortex for proper deployment in the embryo, probably through reorganization of the actin cytoskeleton. These findings uncover a surprising parallel between flies and frogs, as cortically tethered Xenopus Vg1 mRNA undergoes a similar redistribution during oocyte maturation [7]. Our results thus highlight a conserved mechanism for regulating mRNA anchoring and redeployment during the oocyte-to-embryo transition.
2006

Abstract
Patterning of the anterior-posterior body axis of the Drosophila embryo requires production of Nanos protein selectively in the posterior. Spatially restricted Nanos synthesis is accomplished by translational repression of unlocalized nanos mRNA together with translational activation of posteriorly localized nanos. Repression of unlocalized nanos mRNA is mediated by a bipartite translational control element (TCE) in its 3' untranslated region. TCE stem-loop II functions during embryogenesis, through its interaction with the Smaug repressor. Stem-loop III represses unlocalized nanos mRNA during oogenesis, but trans-acting factors that carry out this function have remained elusive. Here we identify a Drosophila hnRNP, Glorund, that interacts specifically with stem-loop III. We establish that the ability of the TCE to repress translation in vivo reflects its ability to bind Glorund in vitro. These data, together with the analysis of a glorund null mutant, reveal a specific role for an hnRNP in repression of nanos translation during oogenesis.

